import os
import h5py
import numpy as np
import pandas as pd
import geopandas as gp
from shapely.geometry import Point
import geoviews as gv
from geoviews import opts, tile_sources as gvts
import holoviews as hv
gv.extension('bokeh', 'matplotlib')
The Global Ecosystem Dynamics Investigation (GEDI) mission aims to characterize ecosystem structure and dynamics to enable radically improved quantification and understanding of the Earth's carbon cycle and biodiversity. The GEDI instrument produces high resolution laser ranging observations of the 3-dimensional structure of the Earth. GEDI is attached to the International Space Station and collects data globally between 51.6$^{o}$ N and 51.6$^{o}$ S latitudes at the highest resolution and densest sampling of any light detection and ranging (lidar) instrument in orbit to date. The Land Processes Distributed Active Archive Center (LP DAAC) distributes the GEDI Level 1 and Level 2 products. The L1B and L2 GEDI products are archived and distributed in the HDF-EOS5 file format.
This tutorial was developed using an example use case for a project being completed by the National Park Service. The goal of the project is to use GEDI L2B data to observe tree canopy height, cover, and profile over Redwood National Park in northern California.
This tutorial will show how to use Python to open GEDI L2B files, visualize the full orbit of GEDI points (shots), subset to a region of interest, visualize GEDI canopy height and vertical profile metrics, and export subsets of GEDI science dataset (SDS) layers as GeoJSON files that can be loaded into GIS and/or Remote Sensing software programs.
It is recommended to use Conda, an environment manager to set up a compatible Python environment. Download Conda for your OS here: https://www.anaconda.com/download/. Once you have Conda installed, Follow the instructions below to successfully setup a Python environment on Linux, MacOS, or Windows.
This Python Jupyter Notebook tutorial has been tested using Python version 3.7. Conda was used to create the python environment.
Using your preferred command line interface (command prompt, terminal, cmder, etc.) type the following to successfully create a compatible python environment:
conda create -n geditutorial -c conda-forge --yes python=3.7 h5py shapely geopandas pandas geoviews holoviews
conda activate geditutorial
jupyter notebook
If you do not have jupyter notebook installed, you may need to run:
conda install jupyter notebook
If you prefer to not install Conda, the same setup and dependencies can be achieved by using another package manager such as pip.
This tutorial uses the GEDI L2B observation from June 19, 2019 (orbit 02932). Use the links below to download the files directly from the LP DAAC Data Pool:
The repository containing all of the required files is located at: https://git.earthdata.nasa.gov/projects/LPDUR/repos/gedi-tutorials/browse
import os
import h5py
import numpy as np
import pandas as pd
import geopandas as gp
from shapely.geometry import Point
import geoviews as gv
from geoviews import opts, tile_sources as gvts
import holoviews as hv
gv.extension('bokeh', 'matplotlib')
inDir = os.getcwd() + os.sep # Set input directory to the current working directory
inDir defined above. You will need to download the file directly from the LP DAAC Data Pool in order to execute this tutorial.¶inDir directory defined above.gediFiles = [g for g in os.listdir() if g.startswith('GEDI02_B') and g.endswith('.h5')] # List all GEDI L2B .h5 files in inDir
gediFiles
['GEDI02_B_2019170155833_O02932_T02267_02_001_01.h5']
L2B = 'GEDI02_B_2019170155833_O02932_T02267_02_001_01.h5'
L2B
'GEDI02_B_2019170155833_O02932_T02267_02_001_01.h5'
GEDI02_B: Product Short Name
2019170155833: Julian Date and Time of Acquisition (YYYYDDDHHMMSS)
O02932: Orbit Number
T02267: Track Number
02: Positioning and Pointing Determination System (PPDS) type (00 is predict, 01 rapid, 02 and higher is final)
001: GOC SDS (software) release number
01: Granule Production Version
h5py package.¶gediL2B = h5py.File(L2B, 'r') # Read file using h5py
list(gediL2B.keys())
['BEAM0000', 'BEAM0001', 'BEAM0010', 'BEAM0011', 'BEAM0101', 'BEAM0110', 'BEAM1000', 'BEAM1011', 'METADATA']
list(gediL2B['METADATA'])
['DatasetIdentification']
This contains useful information such as the creation date, PGEVersion, and VersionID. Below, print the file-level metadata attributes.
for g in gediL2B['METADATA']['DatasetIdentification'].attrs: print(g)
PGEVersion VersionID abstract characterSet creationDate credit fileName language originatorOrganizationName purpose shortName spatialRepresentationType status topicCategory uuid
print(gediL2B['METADATA']['DatasetIdentification'].attrs['purpose'])
The purpose of the L2B dataset is to extract biophysical metrics from each GEDI waveform. These metrics are based on the directional gap probability profile derived from the L1B waveform and include canopy cover, Plant Area Index (PAI), Plant Area Volume Density (PAVD) and Foliage Height Diversity (FHD).
beamNames = [g for g in gediL2B.keys() if g.startswith('BEAM')]
beamNames
['BEAM0000', 'BEAM0001', 'BEAM0010', 'BEAM0011', 'BEAM0101', 'BEAM0110', 'BEAM1000', 'BEAM1011']
for g in gediL2B['BEAM0000'].attrs: print(g)
description wp-l2-l2b_githash wp-l2-l2b_version
for b in beamNames:
print(f"{b} is a {gediL2B[b].attrs['description']}")
BEAM0000 is a Coverage beam BEAM0001 is a Coverage beam BEAM0010 is a Coverage beam BEAM0011 is a Coverage beam BEAM0101 is a Full power beam BEAM0110 is a Full power beam BEAM1000 is a Full power beam BEAM1011 is a Full power beam
beamNames = ['BEAM0110']
Note: This step may take a while to complete.
gediL2B_objs = []
gediL2B.visit(gediL2B_objs.append) # Retrieve list of datasets
gediSDS = [o for o in gediL2B_objs if isinstance(gediL2B[o], h5py.Dataset)] # Search for relevant SDS inside data file
[i for i in gediSDS if beamNames[0] in i][0:10] # Print the first 10 datasets for selected beam
['BEAM0110/algorithmrun_flag', 'BEAM0110/ancillary/dz', 'BEAM0110/ancillary/l2a_alg_count', 'BEAM0110/ancillary/maxheight_cuttoff', 'BEAM0110/ancillary/rg_eg_constraint_center_buffer', 'BEAM0110/ancillary/rg_eg_mpfit_max_func_evals', 'BEAM0110/ancillary/rg_eg_mpfit_maxiters', 'BEAM0110/ancillary/rg_eg_mpfit_tolerance', 'BEAM0110/ancillary/signal_search_buff', 'BEAM0110/ancillary/tx_noise_stddev_multiplier']
pandas dataframe.¶lonSample, latSample, shotSample, qualitySample, beamSample = [], [], [], [], [] # Set up lists to store data
# Open the SDS
lats = gediL2B[f'{beamNames[0]}/geolocation/lat_lowestmode'][()]
lons = gediL2B[f'{beamNames[0]}/geolocation/lon_lowestmode'][()]
shots = gediL2B[f'{beamNames[0]}/geolocation/shot_number'][()]
quality = gediL2B[f'{beamNames[0]}/l2b_quality_flag'][()]
# Take every 100th shot and append to list
for i in range(len(shots)):
if i % 100 == 0:
shotSample.append(str(shots[i]))
lonSample.append(lons[i])
latSample.append(lats[i])
qualitySample.append(quality[i])
beamSample.append(beamNames[0])
# Write all of the sample shots to a dataframe
latslons = pd.DataFrame({'Beam': beamSample, 'Shot Number': shotSample, 'Longitude': lonSample, 'Latitude': latSample,
'Quality Flag': qualitySample})
latslons
| Beam | Shot Number | Longitude | Latitude | Quality Flag | |
|---|---|---|---|---|---|
| 0 | BEAM0110 | 29320618800000001 | 111.996300 | -51.803868 | 0 |
| 1 | BEAM0110 | 29320604600000101 | 112.039132 | -51.803905 | 0 |
| 2 | BEAM0110 | 29320614600000201 | 112.080271 | -51.803836 | 0 |
| 3 | BEAM0110 | 29320600400000301 | 112.121445 | -51.803737 | 0 |
| 4 | BEAM0110 | 29320610400000401 | 112.162622 | -51.803621 | 0 |
| ... | ... | ... | ... | ... | ... |
| 9792 | BEAM0110 | 29320617400979201 | 88.208452 | -51.803578 | 0 |
| 9793 | BEAM0110 | 29320603200979301 | 88.249610 | -51.803614 | 0 |
| 9794 | BEAM0110 | 29320613200979401 | 88.290753 | -51.803581 | 0 |
| 9795 | BEAM0110 | 29320623200979501 | 88.331913 | -51.803548 | 0 |
| 9796 | BEAM0110 | 29320609000979601 | 88.373089 | -51.803506 | 0 |
9797 rows × 5 columns
# Clean up variables that will no longer be needed
del beamSample, quality, qualitySample, gediL2B_objs, latSample, lats, lonSample, lons, shotSample, shots
shapely point generated from each lat/lon location from the shot.¶# Take the lat/lon dataframe and convert each lat/lon to a shapely point
latslons['geometry'] = latslons.apply(lambda row: Point(row.Longitude, row.Latitude), axis=1)
Geopandas GeoDataFrame.¶# Convert to a Geodataframe
latslons = gp.GeoDataFrame(latslons)
latslons = latslons.drop(columns=['Latitude','Longitude'])
latslons['geometry']
0 POINT (111.99630 -51.80387)
1 POINT (112.03913 -51.80391)
2 POINT (112.08027 -51.80384)
3 POINT (112.12145 -51.80374)
4 POINT (112.16262 -51.80362)
...
9792 POINT (88.20845 -51.80358)
9793 POINT (88.24961 -51.80361)
9794 POINT (88.29075 -51.80358)
9795 POINT (88.33191 -51.80355)
9796 POINT (88.37309 -51.80351)
Name: geometry, Length: 9797, dtype: geometry
shapely point below.¶latslons['geometry'][0]
# Define a function for visualizing GEDI points
def pointVisual(features, vdims):
return (gvts.EsriImagery * gv.Points(features, vdims=vdims).options(tools=['hover'], height=500, width=900, size=5,
color='yellow', fontsize={'xticks': 10, 'yticks': 10,
'xlabel':16, 'ylabel': 16}))
redwoodNP = gp.GeoDataFrame.from_file('RedwoodNP.geojson') # Import geojson as GeoDataFrame
redwoodNP
| GIS_LOC_ID | UNIT_CODE | GROUP_CODE | UNIT_NAME | UNIT_TYPE | META_MIDF | LANDS_CODE | DATE_EDIT | GIS_NOTES | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | REDW | None | Redwood | National Park | None | None | None | Shifted 0.06 miles | MULTIPOLYGON (((-124.01829 41.44539, -124.0184... |
redwoodNP['geometry'][0] # Plot GeoDataFrame
# Create a list of geodataframe columns to be included as attributes in the output map
vdims = []
for f in latslons:
if f not in ['geometry']:
vdims.append(f)
vdims
['Beam', 'Shot Number', 'Quality Flag']
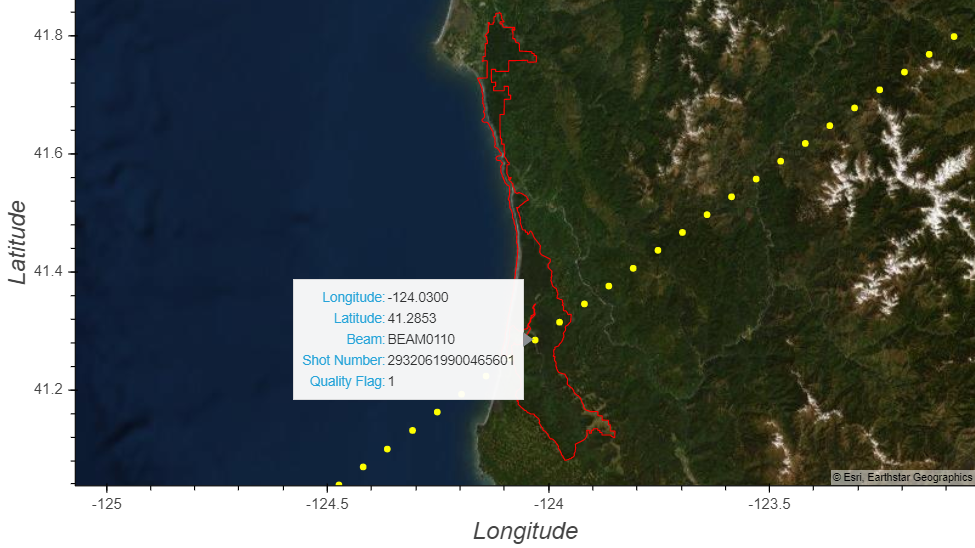
geoviews plots using *) with the point visual mapping function defined above in order to plot (1) the representative GEDI shots, (2) the region of interest, and (3) a basemap layer.¶# Call the function for plotting the GEDI points
gv.Polygons(redwoodNP['geometry']).opts(line_color='red', color=None) * pointVisual(latslons, vdims = vdims)
(HINT: find where the orbit intersects the west coast of the United States)
l2b_quality_flag mean?¶print(f"Quality Flag: {gediL2B[b]['l2b_quality_flag'].attrs['description']}")
Quality Flag: Flag simpilfying selection of most useful data for Level 2B
2932: Orbit Number
06: Beam Number
199: Minor frame number (0-241)
00465601: Shot number within orbit
del latslons # No longer need the geodataframe used to visualize the full GEDI orbit
Detailed product information can be found on the GEDI L2B Product Page.
holoviews.¶len(gediSDS)
1488
beamNames
['BEAM0110']
beamSDS = [g for g in gediSDS if beamNames[0] in g] # Subset to a single beam
len(beamSDS)
186
shot = 29320619500465599
index = np.where(gediL2B[f'{beamNames[0]}/shot_number'][()]==shot)[0][0] # Set the index for the shot identified above
index
465598
pavd = gediL2B[[g for g in beamSDS if g.endswith('/pavd_z')][0]] # PAVD
print(f"Plant Area Volume Density is {pavd.attrs['description']}")
Plant Area Volume Density is Vertical Plant Area Volume Density profile with a vertical step size of dZ
dz layer in order to define the correct vertical step size.¶# Grab vertical step size
dz = gediL2B[f'{beamNames[0]}/ancillary/dz'][0]
dz
5.0
print(f"The shape of PAVD is {pavd.shape}.")
The shape of PAVD is (979699, 30).
# Bring in the desired SDS
elev = gediL2B[f'{beamNames[0]}/geolocation/elev_lowestmode'][()] # Latitude
lats = gediL2B[f'{beamNames[0]}/geolocation/lat_lowestmode'][()] # Latitude
lons = gediL2B[f'{beamNames[0]}/geolocation/lon_lowestmode'][()] # Longitude
shotElev = elev[index]
shotLat = lats[index]
shotLon = lons[index]
shotPAVD = pavd[index]
print(f"The shot is located at: {str(shotLat)}, {str(shotLon)} (shot ID: {shot}, index {index}) and is from {beamNames[0]}.")
The shot is located at: 41.28472739326018, -124.03109998658007 (shot ID: 29320619500465599, index 465598) and is from BEAM0110.
pavdAll = []
pavdElev = []
for i, e in enumerate(range(len(shotPAVD))):
if shotPAVD[i] > 0:
pavdElev.append((shot, shotElev + dz * i, shotPAVD[i])) # Append tuple of shot number, elevation, and PAVD
pavdAll.append(pavdElev) # Append to final list
holoviews Path() function, with the PAVD plotted in the third dimension in shades of green.¶path1 = hv.Path(pavdAll, vdims='PAVD').options(color='PAVD', clim=(0,0.13), cmap='Greens', line_width=20, colorbar=True,
width=700, height=550, clabel='PAVD', xlabel='Shot Number',
ylabel='Elevation (m)', fontsize={'title':16, 'xlabel':16, 'ylabel': 16,
'xticks':12, 'yticks':12,
'clabel':12, 'cticks':10})
path1
# Open all of the desired SDS
dem = gediL2B[[g for g in beamSDS if g.endswith('/digital_elevation_model')][0]][()]
zElevation = gediL2B[[g for g in beamSDS if g.endswith('/elev_lowestmode')][0]][()]
zHigh = gediL2B[[g for g in beamSDS if g.endswith('/elev_highestreturn')][0]][()]
zLat = gediL2B[[g for g in beamSDS if g.endswith('/lat_lowestmode')][0]][()]
zLon = gediL2B[[g for g in beamSDS if g.endswith('/lon_lowestmode')][0]][()]
canopyHeight = gediL2B[[g for g in beamSDS if g.endswith('/rh100')][0]][()]
quality = gediL2B[[g for g in beamSDS if g.endswith('/l2b_quality_flag')][0]][()]
degrade = gediL2B[[g for g in beamSDS if g.endswith('/degrade_flag')][0]][()]
sensitivity = gediL2B[[g for g in beamSDS if g.endswith('/sensitivity')][0]][()]
pavd = gediL2B[f'{beamNames[0]}/pavd_z'][()]
shotNums = gediL2B[f'{beamNames[0]}/shot_number'][()]
# Create a shot index
shotIndex = np.arange(shotNums.size)
canopyHeight = canopyHeight / 100 # Convert RH100 from cm to m
print(f"The shape of Canopy Height is {canopyHeight.shape} vs. the shape of PAVD, which is {pavd.shape}.")
The shape of Canopy Height is (979699,) vs. the shape of PAVD, which is (979699, 30).
# Set up an empty list to append to
pavdA = []
for i in range(len(pavd)):
# If any of the values are fill value, set to nan
pavdF = [np.nan]
for p in range(len(pavd[i])):
if pavd[i][p]!= -9999:
pavdF.append(pavd[i][p]) # If the value is not fill value, append to list
pavdA.append(pavdF) # Append back to master list
len(pavdA)
979699
# Take the DEM, GEDI-produced Elevation, and Canopy height and add to a Pandas dataframe
transectDF = pd.DataFrame({'Shot Index': shotIndex, 'Shot Number': shotNums, 'Latitude': zLat, 'Longitude': zLon,
'Tandem-X DEM': dem, 'Elevation (m)': zElevation, 'Canopy Elevation (m)': zHigh,
'Canopy Height (rh100)': canopyHeight, 'Quality Flag': quality, 'Degrade Flag': degrade,
'Plant Area Volume Density': pavdA, 'Sensitivity': sensitivity})
transectDF
| Shot Index | Shot Number | Latitude | Longitude | Tandem-X DEM | Elevation (m) | Canopy Elevation (m) | Canopy Height (rh100) | Quality Flag | Degrade Flag | Plant Area Volume Density | Sensitivity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 29320618800000001 | -51.803868 | 111.996300 | -999999.0 | 21242.515625 | 21242.515625 | 0.0 | 0 | 0 | [nan] | -3.436965 |
| 1 | 1 | 29320618900000002 | -51.803867 | 111.996712 | -999999.0 | 21242.505859 | 21242.505859 | 0.0 | 0 | 0 | [nan] | 30.496670 |
| 2 | 2 | 29320619000000003 | -51.803867 | 111.997123 | -999999.0 | 21242.496094 | 21242.496094 | 0.0 | 0 | 0 | [nan] | 8.071431 |
| 3 | 3 | 29320619100000004 | -51.803867 | 111.997535 | -999999.0 | 21242.484375 | 21242.484375 | 0.0 | 0 | 0 | [nan] | -212.896439 |
| 4 | 4 | 29320619200000005 | -51.803866 | 111.997946 | -999999.0 | 21242.474609 | 21242.474609 | 0.0 | 0 | 0 | [nan] | -6.853874 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 979694 | 979694 | 29320618400979695 | -51.803445 | 88.411747 | -999999.0 | 18017.906250 | 18017.906250 | 0.0 | 0 | 0 | [nan] | 22.138037 |
| 979695 | 979695 | 29320618500979696 | -51.803445 | 88.412159 | -999999.0 | 18017.296875 | 18017.296875 | 0.0 | 0 | 0 | [nan] | 4.475757 |
| 979696 | 979696 | 29320618600979697 | -51.803444 | 88.412570 | -999999.0 | 18017.884766 | 18017.884766 | 0.0 | 0 | 0 | [nan] | 10.112548 |
| 979697 | 979697 | 29320618700979698 | -51.803444 | 88.412981 | -999999.0 | 18017.275391 | 18017.275391 | 0.0 | 0 | 0 | [nan] | 424.691803 |
| 979698 | 979698 | 29320618800979699 | -51.803443 | 88.413393 | -999999.0 | 18017.263672 | 18017.263672 | 0.0 | 0 | 0 | [nan] | 15.887813 |
979699 rows × 12 columns
# Plot Canopy Height
canopyVis = hv.Scatter((transectDF['Shot Index'], transectDF['Canopy Height (rh100)']))
canopyVis.opts(color='darkgreen', height=500, width=900, title=f'GEDI L2B Full Transect {beamNames[0]}',
fontsize={'title':16, 'xlabel':16, 'ylabel': 16}, size=0.1, xlabel='Shot Index', ylabel='Canopy Height (m)')